IRIS
Background
In recent years, the advances of transcriptomics technologies have allowed us to perform gene expression profiling on many tissue locations with spatial localization information1, enabling the characterization of the transcriptomic landscape on complex tissues.
Overview of IRIS
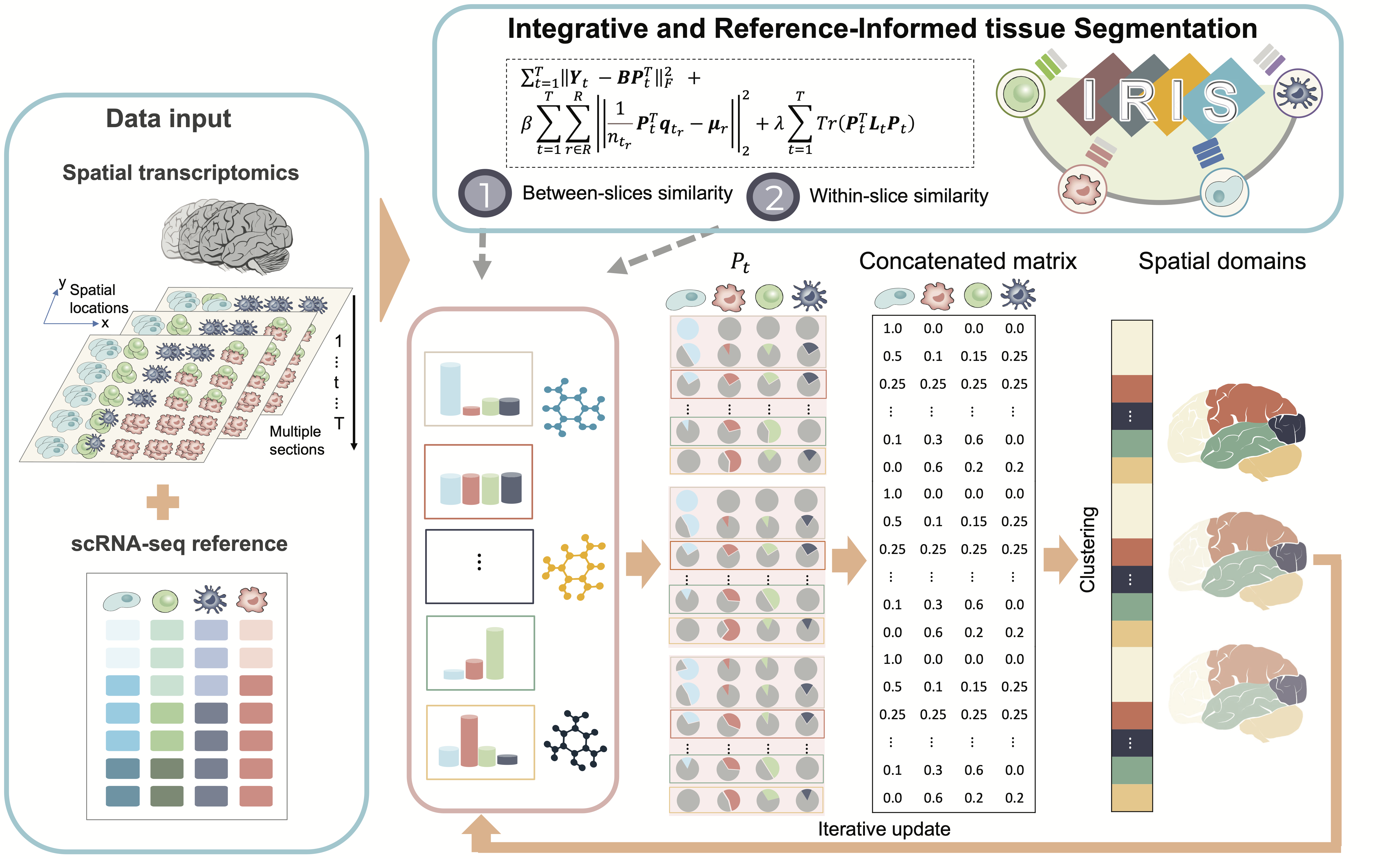
we have developed a novel computational method, Integrative and Reference-Informed tissue Segmentation (IRIS), for spatial domain detection. IRIS is accurate, scalable, and robust, showcasing its advantages across six SRT datasets from different tissues and species, covering 37 tissue slices sequenced using 10x Visium, ST, Slide-seq, Vizgen MERFISH, Stereo-seq, and 10x Xenium. Our results show that IRIS considerably surpassed existing methods in accuracy for spatial domain detection with significantly greater computational efficiency. The unparalleled accuracy and computational gains delivered by IRIS make it an indispensable tool for integrated tissue segmentation in SRT studies (Figure 1).
Paper
Accurate and Efficient Integrative Reference-Informed Spatial Domain Detection for Spatial Transcriptomics
Paper Published in Nature Methods 2024
Selected important results by IRIS

Tutorial of IRIS
We provided a tutorial for running IRIS in details, with all kinds of visualization plots provided. Package Website: IRIS Tutorial
